-
-
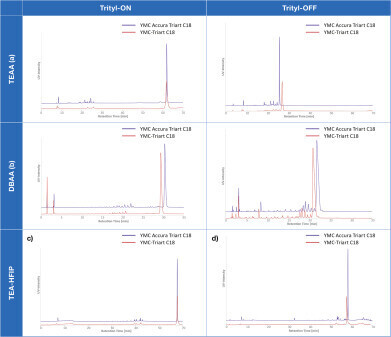 Figure 2: Comparison of stainless-steel and bioinert coated YMC Accura Triart C18 column used for Trityl-ON and Trityl-OFF.
Figure 2: Comparison of stainless-steel and bioinert coated YMC Accura Triart C18 column used for Trityl-ON and Trityl-OFF. -
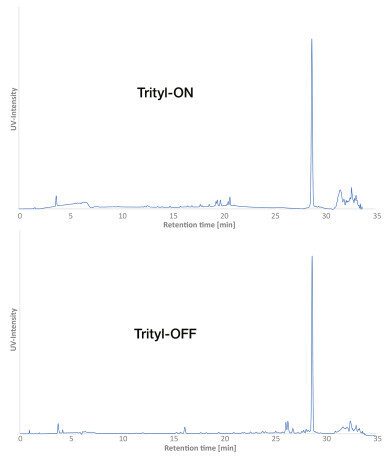 Figure 3: Optimum chromatographic results and conditions for Trityl-ON (top) and Trityl-OFF (bottom).
Figure 3: Optimum chromatographic results and conditions for Trityl-ON (top) and Trityl-OFF (bottom).
Columns (LC)
Optimising the IP-RP analysis of protected and unprotected oligonucleotides
May 21 2024
Short oligonucleotides are usually produced via solid phase synthesis. To achieve a controlled sequence the 5’ terminus is protected with a protecting group such as dimethoxytrityl (DMT). After the successful synthesis, specifically after every synthetic step, the protecting group is removed.
This technical note describes the optimisation of the analysis of two single stranded DNA samples (20mer) using ion pair reversed-phase chromatography (IP-RP). Both samples consist of the same sequence but differ in that the DMT protecting group is still present (Trityl-ON) and the DMT protecting group has been removed (Trityl-OFF). Parameters such as ion pairing agents and their concentration, gradient, column hardware and temperature were optimised for both samples.
Influence of ion pairing agents
Dibutyl ammonium acetate (DBAA), triethyl ammonium acetate (TEAA) and triethylamine (TEA) with 1,1,1,3,3,3-hexafluoride-2-propanol (HFIP) were used for the initial screening at concentrations of 10 mM and 100 mM for HFIP.
When using TEAA, the retention time of the sample is very short. In addition, multiple peaks elute indicating that the ion pairing is incomplete. The chromatogram of the Trityl-ON sample shows that there is no elution when using DBAA. The gradient slope needs to be adjusted. A peak is obtained for Trityl-OFF but its resolution and peak shape are not sufficient. When using TEA-HFIP, both samples elute directly from the column, indicating that the initial conditions are too strong. The gradient and starting conditions were adjusted for further testing.
The optimum concentration of ion pairing agent depends largely on the sample. Therefore, both lower and higher concentrations were tested. The exception was TEAA, which was only tested at a higher concentration of 100 mM, as the sample was not fully paired at a concentration of 10 mM. In addition, different TEA concentrations were tested in combination with 100 mM as well as 200 mM HFIP.
The following concentrations provided the best resolution and peak shape and were therefore selected for further testing:
- 100 mM TEAA
- 10 mM DBAA
- 15 mM TEA + 200 mM HFIP
Influence of column hardware
A direct comparison between regular stainless-steel hardware and YMC Accura column hardware shows the clear advantages of the bioinert column: improved resolution and recovery. The previously optimised ion pairing agent concentrations and gradients were applied. For the further investigations, only the YMC Accura Triart C18 was used.
Conclusions
The following parameters are important for optimising an LC analysis of single-stranded protected/unprotected DNA (Trityl-ON/OFF):
- Choice of ion pairing agent
- Column temperature
- Column hardware
In this case, the best results were achieved with the ion pairing agent TEA-HFIP. Another advantage of these conditions is MS compatibility. However, the high costs of HFIP are a disadvantage. A TEAA buffer is also MS-compatible and offers very good resolution is a cost-effective alternative. Nevertheless, the analysis of DNA using IP-RP must always be individually optimised to the properties of the respective DNA.
It is also important that bioinert column hardware is used when analysing oligonucleotides. Ideally in combination with a bioinert (U)HPLC system. This boosts recoveries and ideal peak shapes are achieved.
Events
Feb 03 2025 Dubai, UAE
Feb 05 2025 Guangzhou, China
Mar 01 2025 Boston, MA, USA
Mar 04 2025 Berlin, Germany
Mar 18 2025 Beijing, China