-
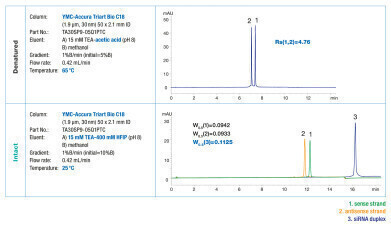 Figure 3: Optimum conditions tested for analysis of a denatured or non-denatured RNA duplex.
Figure 3: Optimum conditions tested for analysis of a denatured or non-denatured RNA duplex. -
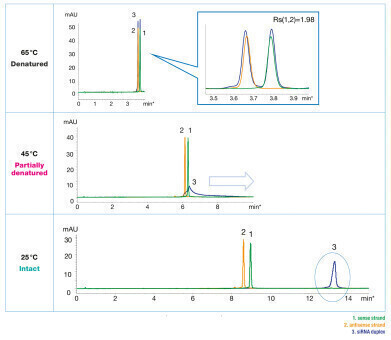 Figure 1: Analysis of the denaturation state of the siRNA duplex at different temperatures.
Figure 1: Analysis of the denaturation state of the siRNA duplex at different temperatures.
Columns (LC)
How to choose the optimum conditions for analysis of siRNA under denaturing and non-denaturing conditions
Aug 22 2023
Small interfering RNAs (siRNAs) usually occur as oligonucleotide duplexes. They can be analysed under non-denaturing conditions so that duplex formation is assured and excessive single-strands are detected. Alternatively, analysis of siRNAs can be conducted under denaturing conditions so that both sense and antisense strands are monitored.
This technical note introduces all relevant aspects of denaturing and non-denaturing ion pair reversed phase high performance liquid chromatography (IP-RP) using a YMC-Accura Triart Bio C18 column. As sample the siRNA duplex targeting the firefly luciferase gene GL2 was used.
Choosing the optimum column temperature
At high temperatures, double-stranded oligonucleotides dissociate. On-column denaturation can be observed by a peak shift since oligonucleotide duplexes adhere stronger to the stationary phase so that single-strands elute earlier.
In figure 1, the different states of the siRNA at different temperatures are visible. At an elevated temperature of 65°C, the double-stranded oligonucleotide is separated into two single-strands. The retention corresponds to the two single-strands analysed separately. At 45°C the siRNA is partially degraded and the retention time increases. At 25°C non-denatured siRNA is detected, which elutes significantly later from the stationary phase.
Selecting the appropriate type and concentration of ion pair reagent
Due to the negatively charged phosphate backbone of siRNAs the retention on hydrophobic stationary phases is low. Therefore, ion pairing agents are needed to neutralise charged siRNAs. This ensures sufficient retention on the stationary phase.
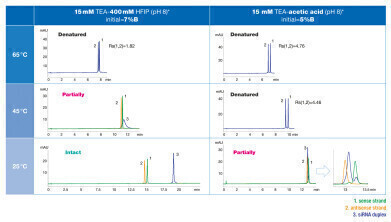
The ion pairing agents n-butylamine (BA), triethylamine (TEA), hexylamine (HA) and dibutylamine (DBA) were tested. The gradient was adjusted individually. At 65°C the siRNA was denatured with all ion pairing agents tested. At 25°C no denaturation was observed. At 45°C the siRNA partially denatured with TEA and HA, and completely denatured with DBA. No denaturation was observed with BA.
Mass spectrometry (MS) applications specifically require volatile ion pairing agents. TEA in combination with 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) as the acidic counter ion and methanol as organic modifier are a common choice.
Therefore, different concentrations of TEA and HFIP were tested. Lower concentrations of TEA and HFIP improved the separation of denatured siRNA marginally. However, the siRNA duplex showed improved peak shape at higher concentrations of TEA and HFIP.
Considering the influence of acetic acid as alternative to HFIP
Besides TEA in combination with HFIP, TEAA buffer (TEA and acetic acid) is commonly used as ion pairing agent. The influence of acetic acid on oligonucleotide separation and denaturation was tested by comparing both ion pairing agents (see figure 2). The use of TEAA resulted in better resolution at 65°C compared to TEA/HFIP. At 25°C siRNA was only partially denatured when TEAA was used whereas it was intact at these conditions tested with TEA/HFIP.
Conclusions
The following factors should be considered during optimisation of siRNA analysis:
- Column temperature
- Choice of ion pair reagent
- Influence of acidic component
The optimum conditions for the tested denaturated and intact siRNA are shown in figure 3.
You will find further information about oligonucleotide analyses in YMC’s Oligonucleotide columns brochure.
Events
May 11 2025 Vienna, Austria
May 18 2025 Tempe. AZ, USA
May 21 2025 Birmingham, UK
Jun 01 2025 Baltimore, MD, USA
Jun 15 2025 Bruges, Belgium